The interferon (IFN) signalling pathway acts as a primary defense against all viruses through the induction of expression of hundreds of genes following infection; the exact functions of each are, at best, poorly understood. In order to gain a better insight into the antiviral mechanism of the induced genes, Schoggins, et al. (2011) performed a sensitive high-throughput screen of the effects of each one on infection with a range of RNA viruses.
 |
| Structure of the IFN-alpha protein. http://www.wikipedia.com/ |
Abstract:
The type I interferon response protects cells against invading viral pathogens. The cellular factors that mediate this defence are the products of interferon-stimulated genes (ISGs). Although hundreds of ISGs have been identified since their discovery more than 25 years ago1, 2, 3, only a few have been characterized with respect to antiviral activity. For most ISG products, little is known about their antiviral potential, their target specificity and their mechanisms of action. Using an overexpression screening approach, here we show that different viruses are targeted by unique sets of ISGs. We find that each viral species is susceptible to multiple antiviral genes, which together encompass a range of inhibitory activities. To conduct the screen, more than 380 human ISGs were tested for their ability to inhibit the replication of several important human and animal viruses, including hepatitis C virus, yellow fever virus, West Nile virus, chikungunya virus, Venezuelan equine encephalitis virus and human immunodeficiency virus type-1. Broadly acting effectors included IRF1, C6orf150 (also known as MB21D1), HPSE, RIG-I (also known as DDX58), MDA5 (also known as IFIH1) and IFITM3, whereas more targeted antiviral specificity was observed with DDX60, IFI44L, IFI6, IFITM2, MAP3K14, MOV10, NAMPT (also known as PBEF1), OASL, RTP4, TREX1 and UNC84B (also known as SUN2). Combined expression of pairs of ISGs showed additive antiviral effects similar to those of moderate type I interferon doses. Mechanistic studies uncovered a common theme of translational inhibition for numerous effectors. Several ISGs, including ADAR, FAM46C, LY6E and MCOLN2, enhanced the replication of certain viruses, highlighting another layer of complexity in the highly pleiotropic type I interferon system.
The interferons are a multifunctional family of around 20 cell-signaling proteins that are secreted from cells following viral infection (as reviewed here). Our cells expend a lot of energy attempting to detect infection and following this, they express high concentrations of IFN proteins. Following secretion, they bind to receptors - and activate - nearby cells alerting them to the viral assault.
Through a complex signaling network a range of genes are actively expressed across the genome that alter the cell in such a way that it becomes harder for viruses to infect them. A rapid antiviral defence system is set-up within the host's tissues and organs. As shown in mice lacking STAT1 a key IFN signal mediator, this IFN signaling network is required to limit viral replication and disease yet also bide time for the development of an adaptive immune response. IFNs are extremely important in our fight against viruses. Only a handful of these IFN-stimulated genes (ISGs) have been characterised while hundreds still sit untouched.
What did they do?
Using previously published gene expression data the group chose 389 ISGs for characterisation. To determine what function these ISGs have on virus replication, Schoggins, et al. developed an intracellular assay in which a retroviral vector expressing high levels of both the individual ISG alongside a red fluorescent protein was used to infect IFN pathway-deficient cells in vitro and then 48 - 72 hours following ISG expression, these exact cells were again infected with a range of RNA viruses expressing a green-fluorescent protein (see figure below). Viruses used included: hepatitis C virus, HIV, yellow fever virus, west Nile virus, Venezuelan equine encephalitis virus and chikungunya virus.
 |
| A) ISG/RFP-expressing retrovirus, B) experimental outline |
 |
| Example results |
Using basic Fluorescent Activated Cell Sorting (FACs) they were able to sort the cells based upon what colour they were, for example: red and the ISG inhibited the virus; green and it did not. More importantly they were able to specifically quantify the levels of green and red to assess the exact levels of virus replication in the cell population. The strongest inhibitors were extensively validated. On top of being expressed individually, some ISGs were expressed in combination to determine whether their antiviral effects were additive. The group were finally able to pinpoint where in the virus replication cycle inhibition took place: entry, transcription, translation, replication or exit.
The major findings included:
- Most ISGs inhibit virus replication
As would be expected for an antiviral response, and one which has been shown to already inhibit a wide range of viruses, the majority of the tested ISGs inhibited replication to some degree across a range of viruses looked at. This is a good thing. This means your assay is doing what is supposed to do.
- There are two types of antiviral genes: modest and strong inhibitors
Owing to how the IFN system has evolved, we may categorise the ISGs into two antiviral classes: modest and strong inhibitors. The modest ones act specifically, targeting limited aspects of the virus replication cycle; the stronger ones function as a positive feedback, increasing the expression of key IFN signaling genes
- Inhibition is additive - more ISGs equals more inhibition
Upon the activation of the IFN pathway, hundreds of genes are upregulated resulting in an antiviral state within the cell. The defence system is not set up so that protection lies in the hands of a single gene/protein but in the hands of many - it is a truly cooperative process. In this system, following the expression of different combinations of ISGs together within a single cell, the author's noted that inhibition of viral replication increased.
- Translational inhibition is the most common antiviral mechanism
The group asked, using a number of assays, at what point do these ISG's inhibit hepatitis C virus replication - is it: entry or translation of HCV mRNA/genome. They found that, in this case, it was not entry but translation. Although, this effect is probably particular to positive-sense RNA viruses.
- Some ISGs enhanced virus infection
Interestingly, they found that following expression of a number of ISG's, virus replication increased. Something of a surprise for an antiviral pathway. The mechanisms of why/how these genes did this was not addressed but we can assume that in a real-life infection, the other 380-odd inhibitory genes also upregulated would cancel these out.
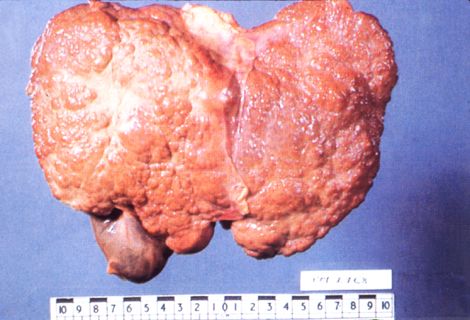 |
| HCV-induced hepatocellular carcinoma. WIll this IFN research aid in potential HCV antivirals? http://www.stanford.edu |
Some caveats exist, however:
- Over-expression systems may not reflect in vivo situation
The results from this work are extremely interesting, important and useful yet the way it was done may not accurately represent what happens when you are infected. Although, I feel that this was not what was set out to be determined by the investigators, which I think were stimulated by the need of novel antiviral candidates. This research will of course aid in that field through the identification of potentially life-saving targets.
- Only a relatively small number of diverse viruses were screened
The viruses tested in this system included only a relatively small sample of virus diversity - mainly focusing upon small positive sense RNA viruses (HCV etc.), and one retrovirus: HIV-1. We saw that a number of these ISG's inhibited these viruses but does this reflect what may happen with other viruses? What effect would they have on large DNA viruses, negative-sense RNA viruses or double-stranded RNA viruses? While the fact that the majority of the genes inhibited replication probably wouldn't change, the specifics most likely would. A number of these genes would specifically target pathways and systems that are preferentially used by these small positive sense RNA viruses while some would target those used by other viruses.This experiment would only 'see' those affecting these small positive sense RNA viruses. Although, this may not be a bad thing. These viruses are major causes of morbidity and mortality in both human and other animal populations worldwide.
- Doesn't take into consideration virus IFN modulation
Every virus probably has different ways of inhibited the IFN response themselves before they are eliminated from the host following ISG expression. This system would not pick up of the majority of these modulatory mechanisms as they occur prior to the expression of ISGs. So, again, as I mentioned before, this work does not reflect what would happen during infection in vivo and nor does it really matter.
Schoggins, J., Wilson, S., Panis, M., Murphy, M., Jones, C., Bieniasz, P., & Rice, C. (2011). A diverse range of gene products are effectors of the type I interferon antiviral response Nature DOI: 10.1038/nature09907

















No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS