Have you ever wanted to visualise viral infection? Ever wanted to observe how they enter and spread throughout their host organism? Ever wanted to know how exactly they caused disease - at the cellular and whole-organism level? Well, this may be entirely possible using fluorescent-labeled recombinant viruses infecting a relevant model system.
[caption id="" align="aligncenter" width="504" caption="GFP-virus infected cells"]

[/caption]
So how does it work?Lemon et al recently report the continued investigation of measles virus pathogenesis in a non-human primate (
Macaque) model utilising a green-fluorescent protein (
GFP) expressing virus. Upon infection of host cells, viral transcription leads to the very high expression of GFP, flooding the cytoplasm with this fluorescent ‘tag’. Subsequent
microscopy, imaging and immunohistochemistry allows for the identification and location of the infected cells, tissues and organs - see image above. Tracking of cellular infection allows us to decipher the development of MeV entry, spread and replication at both the cellular and whole-organism level throughout the entire infection. Studies such as these give an unprecedented view of viral infection in a means directed related to that of human infection. This model even allows for macroscopic real-time detection of fluorescence and hence viral infection.
Why is this important for measles?Despite a highly effective vaccine and significant global control initiatives,
measles infection still accounts for significant morbidity and mortality worldwide, mostly in the developing world (164,00 deaths in 2008). This is mostly attributable to the profound immunosuppression induced allowing for further infection with opportunistic pathogens.
Currently, much is known about measles pathogenesis yet the molecular mechanisms of such are poorly understood and it is therefore of great interest to better understand these processes by which MeV infects and causes disease in humans. Knowledge of such may facilitate the development of more
effective and safer vaccines for measles and indeed other viral pathogens.
Viruses being obligate intra-cellular parasites, must enter and exit cells in order to survive. Most of viral pathogenesis can therefore be attributed to the effects of viral replication of host cells and tissues; a major determinant of which is the expression of receptors on host cells surfaces allowing viral entry, infection and replication. Currently only a single receptor – CD150 - (otherwise known as signalling lymphocyte activation molecule SLAM) has been discovered that wild-type pathogenic MeV uses to enter host cells; the distribution of which only explains part of measles pathogenesis as epithelial and neuronal cells (important target cells) do not express the protein. As indicated by this receptor being expressed on lymphocytes and other immune cells, MeV is a highly lymphotropic virus! But if epithelial cells fail to express the receptor on their surface, how come its possible for MeV to enter via these cells?
The classical view of measles pathogenesis was that free-virus entered the host through the respiratory route, infecting and primarily replicating within the epithelial cell lining of the respiratory tract. Newly produced virus spreads to nearby lymph nodes where infected monocytes – a type of immune cell - facilitates viral dissemination throughout the host, resulting in the well-known symptoms of measles. The problem with this being that epithelial cells and unstimulated monocytes fail to express the MeV receptor CD150 and infection should therefore not occur. Recently, it has been shown (again using a GFP expressing virus in a macaque model) that MeV predominately infects dendritic cells during the peak of infection, ruling out a major role for monocytes. There is also however no direct evidence of MeV primary replication within the epithelium of the respiratory tract at the early stages of infection. So what exactly happens during the start of infection and does it develop? GFP-expressing viruses may shed light on this question.
[caption id="" align="aligncenter" width="415" caption="Diagramatic representation of the cellular composition of the human respiratory tract - notice the epithelial cell lining and the alveolar macrophages. Dendritic cells are however not shown on this diagram."]
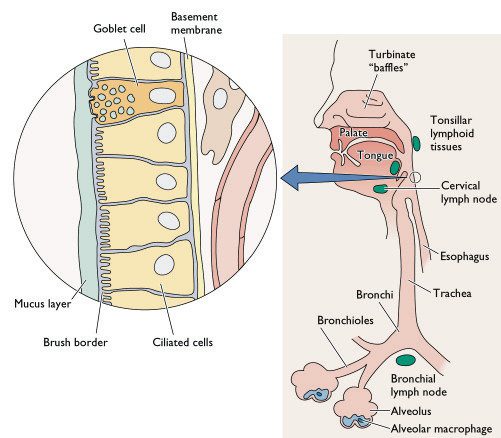
[/caption]
So how can we study the early stages of infection?The incubation period of measles is about 2 weeks in humans making it particularly difficult to study the early events of viral infection – the kind of events like host entry, initial site of replication and subsequent intra-host dissemination - this is where we can use a non-human primate model.
Lemon et al generated a highly virulent recombinant MeV based on viral isolates from an outbreak in Sudan; they engineered the viral genome so that it expressed GFP upon entry into cells – an addition that causes little or no replication defects to the virus. Groups of macaques were subsequently infected via the respiratory route allowing highly sensitive visualisation of GFP expressing cells following necropsy. The early time-points of around 5 days post infection were focussed on in this investigation allowing the determination of the early cell targets - epithelium? Immune cells?
So what did they find?Their results suggest that at the early stages of MeV infection, GFP and hence viral replication is only found in immune cells within the respiratory tract and not the epithelial lining.
Dendritic cells and alveolar macrophages are believed to capture viral particles in the lungs allowing spread via infected cells. This is known as a Trojan horse entry mechanism like that used by HIV to pass through mucosal tissues and infect humans - see below. This infection allows for spread and localised replication within nearby lymphoid tissues and then on to draining lymph nodes where massive lymphocyte cell infection may occur facilitating dissemination throughout the host, mainly within lymphoid tissues. Virus can be carried through host blood vessels to other lymphoid target tissues like the tonsils and adenoids and the gut-associated lymphoid tissue ' Peyer's patches'.
[caption id="" align="aligncenter" width="465" caption="HIV entry mechanisms utilising dendritic cells to pass through epithelial cell barriers - the 'Trojan horse' mechanism. This may be directly analogous to MeV entry and primary spread except in the respiratory tract."]

[/caption]
What does this mean?This study clearly demonstrates the importance of non-epithelial cells such as dendritic cells in MeV entry, early replication and subsequent systemic spread. It does not however, rule out a major role for epithelial cells in later stages and in transmission - MeV still infects non-CD150 expressing cells and currently the mechanisms of which are unknown. Focusing on the later stages of infection may allow us to appreciate the other cell targets in pathogenesis and viral transmission. As mentioned previously, the use of fluorescent-labeled viruses offers an unprecedented view of viral entry, spread and pathogenic mechanisms. We should look forward to the time when studies like these are applied to other v
iral and indeed non-viral pathogens.
 Lemon, K., de Vries, R., Mesman, A., McQuaid, S., van Amerongen, G., Yüksel, S., Ludlow, M., Rennick, L., Kuiken, T., Rima, B., Geijtenbeek, T., Osterhaus, A., Duprex, W., & de Swart, R. (2011). Early Target Cells of Measles Virus after Aerosol Infection of Non-Human Primates PLoS Pathogens, 7 (1) DOI: 10.1371/journal.ppat.1001263Coombes, J., & Robey, E. (2010). Dynamic imaging of host–pathogen interactions in vivo Nature Reviews Immunology, 10 (5), 353-364 DOI: 10.1038/nri2746
Lemon, K., de Vries, R., Mesman, A., McQuaid, S., van Amerongen, G., Yüksel, S., Ludlow, M., Rennick, L., Kuiken, T., Rima, B., Geijtenbeek, T., Osterhaus, A., Duprex, W., & de Swart, R. (2011). Early Target Cells of Measles Virus after Aerosol Infection of Non-Human Primates PLoS Pathogens, 7 (1) DOI: 10.1371/journal.ppat.1001263Coombes, J., & Robey, E. (2010). Dynamic imaging of host–pathogen interactions in vivo Nature Reviews Immunology, 10 (5), 353-364 DOI: 10.1038/nri2746


































 [/caption]
[/caption] [/caption]
[/caption] [/caption]
[/caption]